Feature Subset Selection in r using Embedded Approach
Regularization is a regression technique that shrinks feature coefficients towards zero to simplify the learning model, reduce overfitting while promising the least amount of error on new unknown data.
Ranking measures the importance of individual features. Filters measure the significance of feature subsets to both classifiers, as well as, between features within a subset. Wrappers iterate through potential subsets scoring each one using a learning algorithm.
The fourth approach, embedded feature selection, performs feature selection (or, as we will see, feature modification) during model training and uses the resulting performance as a guide for selection.
To understand the effectiveness of an embedded feature selection technique we must first understand how to evaluate the performance of a trained model.

Evaluating a Model
One of the fundamental assumptions behind supervised machine learning is that there exists some true target function f(x) that will precisely predict y using some unknown data. This true function is unknowable.
The goal, then, is to find a usable function which approximates the true function. We do this by trying various models and measuring the errors between known values and the predictions being made by the model.
Another fundamental assumption is that the number of data instances provided for such training is fixed. Since some date is needed to train the model and some different data to validate the approximation we need to split the known data into two separate groups. Different splits produce different results.
A technique called cross-validation performs a split and allows the validation to be run a number of times. The data is broken up into some number of equal parts (a common number is ten) and Part 1 is used to train the model and produces the training error. Parts 2-10 are then used to validate the model, by running each set across the model separately. These nine error sets are then averaged and result in the cross-validation error.
In-sample Error
Both of these errors result from using known (sample) data and are called in-sample errors. If we plot these two errors as a function of the number of data instances we gain insight into the performance of the model being trained. These plots are called learning curves.
Once type of learning curve shows the relationship between the cross-validation error and the training error.
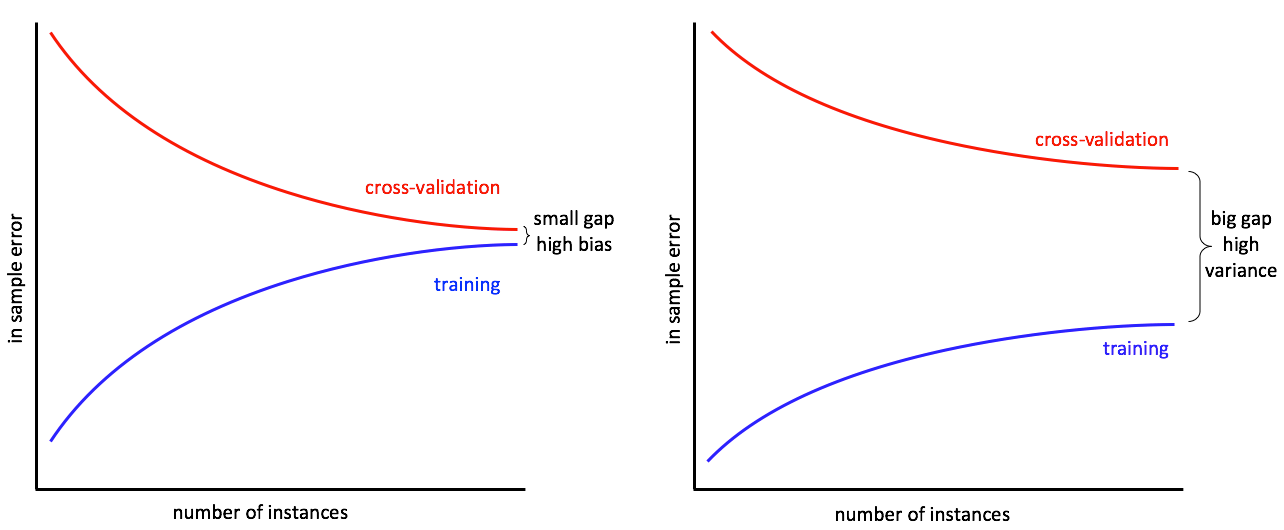
The gap between training and cross-validation errors represents the two competing properties of statistical learning models: bias and variance.
Bias is a measurement of how well a model approximates the true function. If the approximating (trained) model is much simpler then the actual true function then the bias is high. The goal in training is to minimize the bias.
Variance is the difference between predictions over different sets of data. If a model has high variance then even small changes in the training data can result in large changes in the results. The goal is to minimize the difference in outputs (variance) over different sets of data.
The goal of machine learning is to balance model complexity while minimizing error. In the graphic below the ideal complexity is the dashed vertical line.
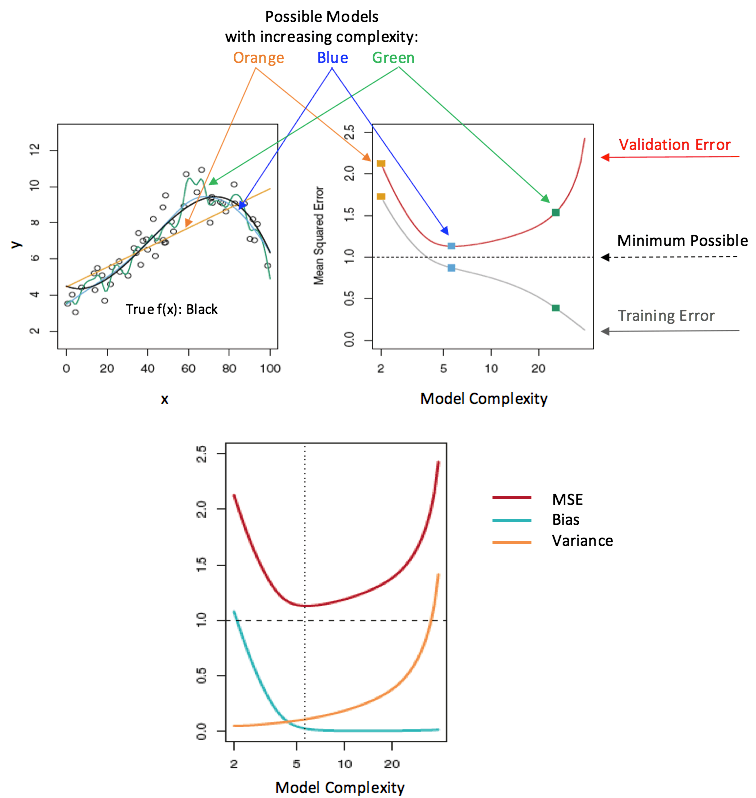
In the illustration above MSE is the mean squared error which is the difference between the training and validation errors. The smaller the MSE the better the model.
Simple Example
Imagine you want to predict a persons salary. Say you trained the model using just a person’s age as a feature. This would be a simple, non-complex, model. This model would be said to have high bias, also known as an underfitted model.
If you then added features: sex, education, profession, model of car, years of experience, parents profession, weight and height the model is more complex. During training, the model tries to account for every feature. This results in high variance or overfitting.
One way of controlling overfitting is by adding an extra term to the MSE calculation during training to penalize large coefficients in the model. One approach adds a term to penalize the sum of the squared coefficients, known as Ridge Regression or L2 regularization. Another technique adds a term to penalize the sum of the absolute value of the coefficients, known as the Lasso or L1 regularization.
Out-of-sample Error
In-sample error, produced during training and validation, is based on known data. But the end goal is to minimize out-of-sample error E(out), the error produced when predicting y using unseen data.
It is possible to show that E(out) can be decomposed into the following components:\(\)$$Predicted\;E(out)\;\; = \;\;Bias^2 \;\;+\;\; Variance\;\; +\;\; Irreducible\ Error$$$$E(out)\;\; =\;\; ( E\ [\hat f(x) ]\; –\; f(x) )^2 \;\;\; + \;\;\; E\ [(\hat f(x)^2] \ \ (E[\hat f(x)])^2 \;\;\; + \;\;\; σ_e^2$$
Where the irreducible error is some amount of immeasurable noise in the data. To minimize E(out), during training, the model complexity is varied until both the bias and variance are minimized.
One way to reduce model complexity is to simply remove redundant, noisy features, or features that are correlated with each other or uncorrelated to the class you are trying to predict. A second technique is called regularization where the coefficients of the features are adjusted towards zero which has the effect of reducing feature strength. Regularization modifies the complexity of the model.
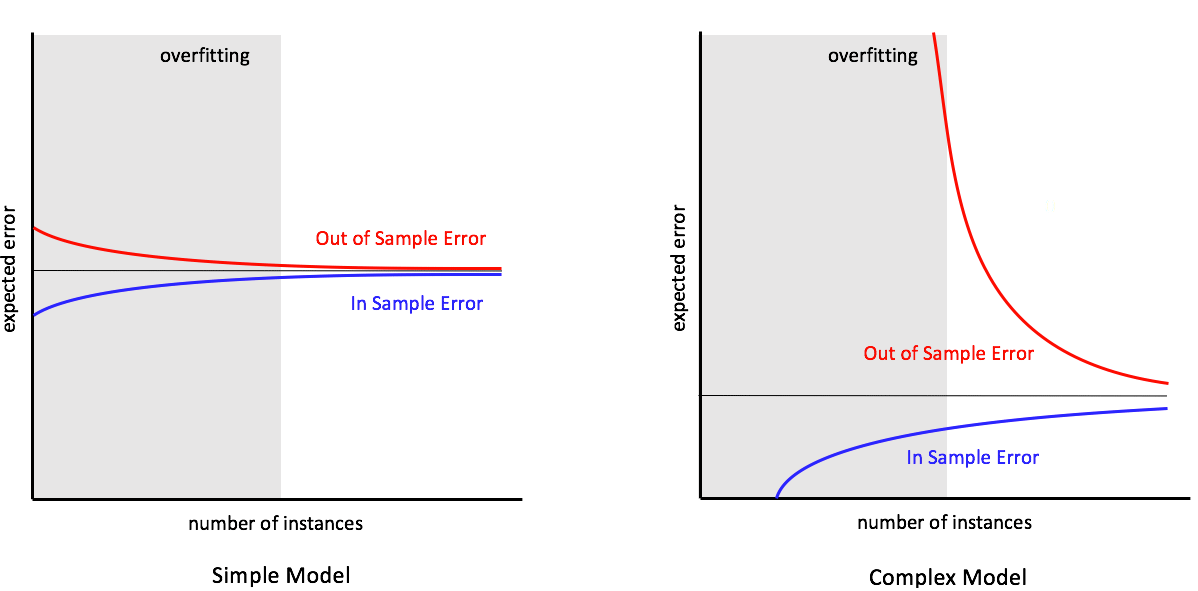
“Match the model complexity to the data resources not to the target complexity” Yaser Abu-Mostafa
Embedded Approaches to Feature Selection
The previous approaches at the top of the post remove features completely. The following approaches use a technique called regularization which uses a penalty term to adjust the coefficients of the features towards zero. In r the results of glmnet’s cross validation contain a number of results. One is lambda.1se which is the largest value of lambda where the error is within one standard error of the minimum and the one we will use below to plot the best coefficients.
Ridge Regression
We’ll use the same data set as previous posts. Ridge methods involve adding a tuning parameter λ to the model, which is designed to impose a penalty on each term’s coefficient based on λ’s size. If λ = 0 the penalty has no effect and the model produces the least squares estimates similar to linear regression. However when λ → ∞ the penalty pushes the coefficient estimates to zero.
Ridge regression is also known as L2 regularization which is the penalizing of the squared magnitude (coefficients) of all features. L2 tends to penalize and therefore shrink larger feature coefficients more than the lasso regularization.
Because each value of λ (lambda) will produce a different set of coefficients it is important to choose the right lambda which glmnet does during cross validation. The cross validation result contains multiple values. One is the value of for lambda (model_name$lambda.1se) which gives the most regularized model with an error within one standard error of the minimum.
library(data.table) # provides enhanced data.frame
library(ggplot2) # plotting
library(glmnet) # ridge, elastic net, and lasso
setwd("/Users/xyz/rData")
oldProducts <- read.csv('existing-product-attributes.csv'
# read data
, sep=","
, header = T
, as.is=T
, stringsAsFactors=F
, check.names = FALSE)
# glmnet requires x matrix (of predictors) and vector (values for y)
x = model.matrix(Volume~.,oldProducts) # matrix of predictors
y = oldProducts$Volume # vector y values
set.seed(123) # replicate results
ridge_model <- cv.glmnet(x, y, alpha=0) # alpha=0 is ridge
best_lambda_ridge <- ridge_model$lambda.1se # largest lambda in 1 SE
ridge_coef <- ridge_model$glmnet.fit$beta[, # retrieve coefficients
ridge_model$glmnet.fit$lambda # at lambda.1se
== best_lambda_ridge]
coef_r = data.table(ridge = ridge_coef) # build table
coef_r[, feature := names(ridge_coef)] # add feature names
to_plot_r = melt(coef_r # label table
, id.vars='feature'
, variable.name = 'model'
, value.name = 'coefficient')
ggplot(data=to_plot_r, # plot coefficients
aes(x=feature, y=coefficient, fill=model)) +
coord_flip() +
geom_bar(stat='identity', fill='brown4', color='blue') +
facet_wrap(~ model) + guides(fill=FALSE)
The Lasso
Lasso is also called L1 regularization which is used when the data is noisy. Unlike ridge regression, lasso can effectively remove features as it sets their coefficients to zero. Removing terms in this way is sometimes referred to as finding the sparsest solution.
library(data.table) # provides enhanced data.frame
library(ggplot2) # plotting
library(glmnet) # ridge, elastic net, and lasso
setwd("/Users/xyz/rData")
oldProducts <- read.csv('existing-product-attributes.csv'
# read data
, sep=","
, header = T
, as.is=T
, stringsAsFactors=F
, check.names = FALSE)
# glmnet requires x matrix (of predictors) and vector (values for y)
x = model.matrix(Volume~.,oldProducts) # matrix of predictors
y = oldProducts$Volume # vector y values
set.seed(123) # replicate results
lasso_model <- cv.glmnet(x, y, alpha=1) # alpha=1 is lasso
best_lambda_lasso <- lasso_model$lambda.1se # largest lambda in 1 SE
lasso_coef <- lasso_model$glmnet.fit$beta[, # retrieve coefficients
lasso_model$glmnet.fit$lambda # at lambda.1se
== best_lambda_lasso]
coef_l = data.table(lasso = lasso_coef) # build table
coef_l[, feature := names(lasso_coef)] # add feature names
to_plot_r = melt(coef_l # label table
, id.vars='feature'
, variable.name = 'model'
, value.name = 'coefficient')
ggplot(data=to_plot_l, # plot coefficients
aes(x=feature, y=coefficient, fill=model)) +
coord_flip() +
geom_bar(stat='identity', fill='brown4', color='blue') +
facet_wrap(~ model) + guides(fill=FALSE)
Elastic Net
Ridge regression performs better than ordinary least squares (OLS) but it does not result in a parsimonious model; however, the lasso also has some problems.
There are three scenarios to consider when choosing regularization techniques based on shrinkage [Zou, Hastie].
- There are more predictors than instances of data which limits accuracy of the lasso’s feature selection.
- There is high correlation between pairs of features. Lasso selects only one indiscriminately.
- There are more instances than predictors. Ridge outperforms lasso but does not remove features.
The elastic net is a combination of ridge and lasso regularization. Like lasso, it does feature selection and shrinkage of the coefficients but it also selects groups of correlated features. Like an elastic fishing net it retains ‘all the big fish’ and can often outperform the lasso’s prediction accuracy. Elastic net solves the problem the lasso has in the first two scenarios and delivers better accuracy than lasso in the third.
library(data.table) # provides enhanced data.frame
library(ggplot2) # plotting
library(glmnet) # ridge, elastic net, and lasso
setwd("/Users/xyz/rData")
oldProducts <- read.csv('existing-product-attributes.csv'
# read data
, sep=","
, header = T
, as.is=T
, stringsAsFactors=F
, check.names = FALSE)
# glmnet requires x matrix (of predictors) and vector (values for y)
x = model.matrix(Volume~.,oldProducts) # matrix of predictors
y = oldProducts$Volume # vector y values
set.seed(123) # replicate results
en_model <- cv.glmnet(x, y, alpha=0) # 0 < alpha < 1 elastic net
best_lambda_ridge <- en_model$lambda.1se # largest lambda in 1 SE
en_coef <- en_model$glmnet.fit$beta[, # retrieve coefficients
en_model$glmnet.fit$lambda # at lambda.1se
== best_lambda_en]
coef_en = data.table(elasticNet = en_coef) # build table
coef_en[, feature := names(ridge_coef)] # add feature names
to_plot_r = melt(coef_en # label table
, id.vars='feature'
, variable.name = 'model'
, value.name = 'coefficient')
ggplot(data=to_plot_r, # plot coefficients
aes(x=feature, y=coefficient, fill=model)) +
coord_flip() +
geom_bar(stat='identity', fill='brown4', color='blue') +
facet_wrap(~ model) + guides(fill=FALSE)
Comparing Ridge, Lasso, and Elastic Net Coefficients
By placing all three into a single table we can more easily compare the results of the best coefficients of each approach.
# after running the code above
all_coef = data.table (lasso = lasso_coef,
elastic_net = elastic_net_coef,
ridge = ridge_coef)
# show all coefficients in all three models
all_coef[, feature := names(ridge_coef)]
all_coef
# lasso elastic_net ridge feature
# 0.00000 0.0000000 0.000000e+00 (Intercept)
# 0.00000 0.0000000 1.559799e+01 ProductType
# 0.00000 0.0000000 4.616179e-02 Price
# 3.83083 3.6694915 2.321768e+00 StarReviews5
# 0.00000 1.0736760 4.879834e+00 StarReviews4
# 0.00000 0.0000000 5.247935e+00 StarReviews3
# 0.00000 0.0000000 9.368827e-01 StarReviews2
# 0.00000 0.0000000 -3.974613e-01 StarReviews1
# 0.00000 0.1503263 1.412493e+00 PosServiceReview
# 0.00000 0.0000000 -5.929635e+00 NegServiceReview
# 0.00000 0.0000000 1.863036e+02 WouldRecommend
# 0.00000 0.0000000 9.964489e-03 BestSellersRank
# 0.00000 0.0000000 3.020842e+00 ShippingWeight
# 0.00000 0.0000000 1.475852e+00 ProductDepth
# 0.00000 0.0000000 -1.168473e+01 ProductWidth
# 0.00000 0.0000000 -3.784464e-02 ProductHeight
# 0.00000 0.0000000 -3.574114e+02 ProfitmarginResources
James,Witten, et al. Introduction to Statistical Learning. 2003.
Zou, Hastie. Regularization and variable selection via the elastic net. 2004
Gravio, Chiara. Instrumental variables selection : a comparison between regularization and post-regularization methods. 2015.
Ramakrishnan. What is Ridge Regression in Laymans terms. 2018.
Jain. A Complete Tutorial on Ridge and Lasso Regression in Python. 2016.